The main focus of the Computational Biomedical Imaging Group (CBIG) is on the development of theoretical and algorithmic tools for machine learning in the presence of missing data. CBIG's research applies to biomedical imaging, particularly magnetic resonance imaging (MRI). The research capitalizes on the state-of-the-art dedicated MRI research facilities, high performance computing, and research in MRI and neuroscience at the University of Iowa.
Model-Based Supervised and Unsupervised Deep Learning
Direct inversion schemes that directly recover an image from undersampled measurements are widely used in imaging. By contrast, the model based image image reconstruction framework marries the physics of the acquisition with learnable deep learning modules. The formulation provides a systematic approach for deriving deep architectures for inverse problems with the arbitrary structure. This approach dramatically reduces the demand for training data and training time.
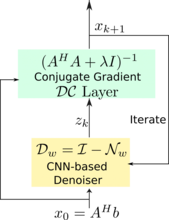
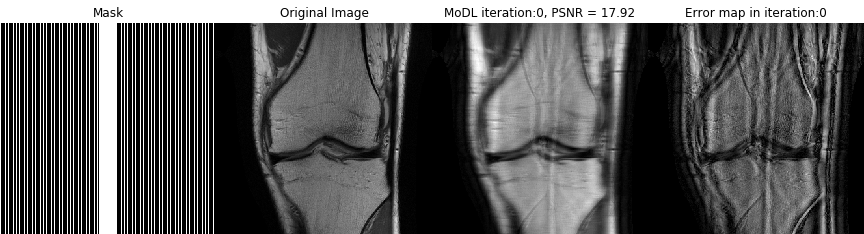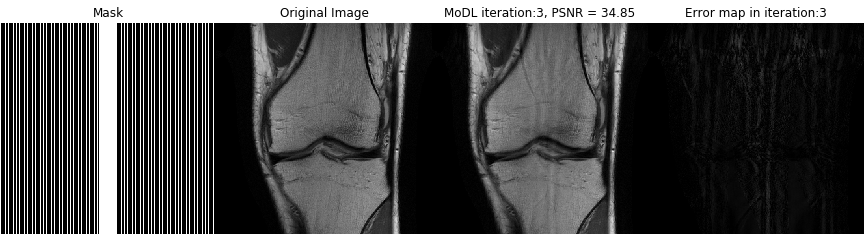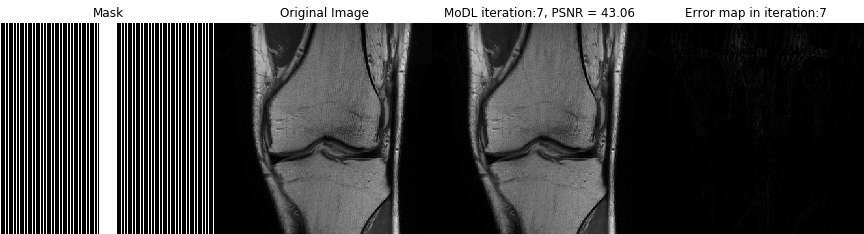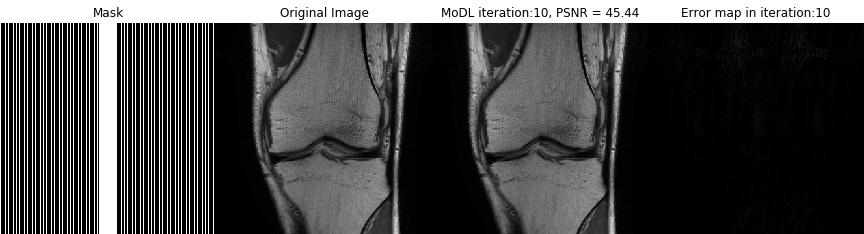
Acquiring fully sampled training data is often challenging in many applications, especially in ultra-high-resolution imaging and dynamic MRI. We introduce novel strategies including ensemble Stein's unbiased risk estimator (ENSURE) to train deep learning problems from undersampled data. Although our focus is on MRI, the approach is broadly applicable to other imaging problems (e.g. deblurring, inpainting), where uncorrupted data is not readily available.
Relevant papers
- MoDL: Model Based Deep Learning Architecture for Inverse Problems
- Dynamic MRI using model-based deep learning and SToRM priors: MoDL-SToRM
- Off-the-grid Model-Based Deep Learning (O-MODL)
- Highly accelerated diffusion MRI using q-MODL
- ENSURE: Ensemble Stein's Unbiased Risk Estimator for Unsupervised Learning
Software
Calibrated parallel MRI with MODL
Smoothness regularization on Manifolds (STORM)
Many subjects in MRI cannot tolerate the breath-held cardiac MRI exams. The STORM framework is a free-breathing and ungated dynamic MRI scheme. It exploits the manifold structure of images in the dataset to recover them from highly undersampled measurements. The data is acquired using a spiral or radial sequence, and the reconstruction relies on a kernel low-rank algorithm.
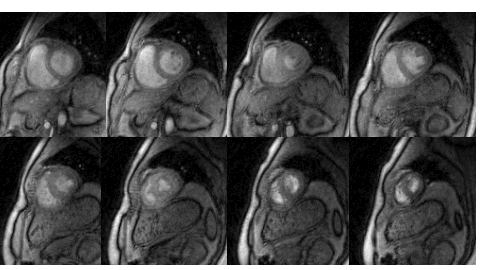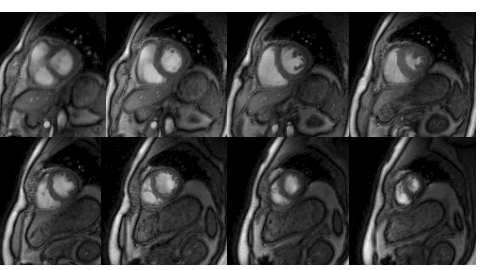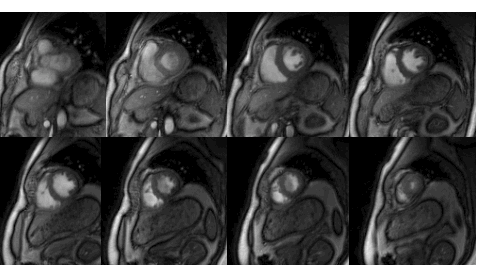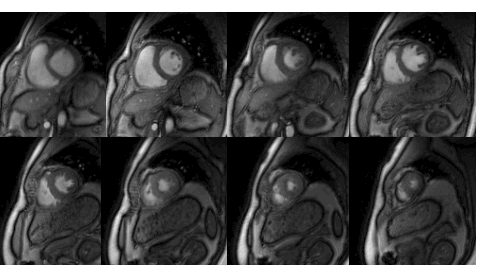
Relevant papers
- Free-breathing and ungated cardiac cine using navigator-less spiral SToRM
- Dynamic MRI using Smoothness Regularization on Manifolds (SToRM)
- Dynamic MRI using model-based deep learning and SToRM priors: MoDL-SToRM,
- Manifold recovery using kernel low-rank regularization: application to dynamic imaging
- Free-breathing & ungated cardiac MRI using iterative SToRM (i-SToRM)
Software:
Deep Structured Low-rank algorithms for uncalibrated MRI
Structured low-rank algorithms (SLR) are efficient in capturing several redundancies in the data that classical compressed sensing (CS) methods fail to capture. For instance, classical multichannel CS methods and MODL require calibration scans to estimate the coil sensitivities. When calibration data is not available or is corrupted by subject motion, these methods offer poor performance. SLR scheme enables the calibration-free recovery of parallel MRI data, offering improved performance over calibration-based strategies, when there is motion errors. However, SLR methods are associated with high computational complexity. Deep SLR approach offers fast image recovery, while offering improved performance over calibration based methods.
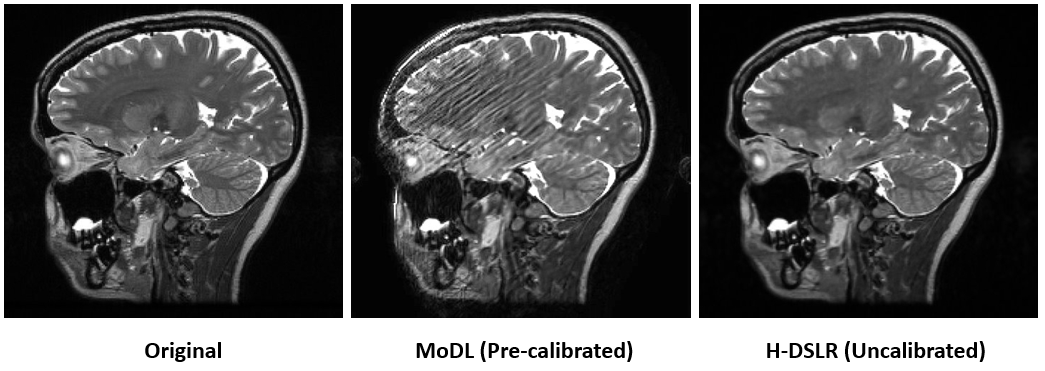
Relevant papers
- Uncalibrated imaging using Deep SLR
- MoDL-MUSSELS: Model-Based Deep Learning for Multi-Shot Sensitivity Encoded Diffusion MRI
Software
Uncalibrated parallel MRI with Deep-SLR
Continuous Domain Compressed Sensing

The theory of compressed sensing (CS) has revolutionized the area of signal processing by overcoming classical resolution limits. However, current discrete CS formulations are often restrictive and result in model mismatch in imaging applications, where the recovery of a continuous domain image from continuous domain measurements is considered. We introduce a novel structured low-rank matrix completion formulation, where the recovery of a continuous domain image from its sparse measurements is considered. In particular, we reformulate sparse recovery of continuous domain signal as a low-rank matrix completion problem in the spectral domain, thus providing the benefit of sparse recovery with performance guarantees. Fast algorithms that are comparable in complexity to discrete compressed sensing methods are also investigated. This framework improved the state of the art in various applications, including dynamic MRI, parametric imaging, and diffuson MRI. For details, see slides of the tutorial at ISBI 2017 by Mathews Jacob and Prof. Jong Chul Ye.
Relevant papers
- Structured Low-Rank Algorithms, Theory, MR Applications, and Links to Machine Learning
- Convex recovery of continuous domain piecewise constant images from non-uniform Fourier samples
- Off-the-Grid Recovery of Piecewise Constant Images from Few Fourier Samples
- A Fast Algorithm for Convolutional Structured Low-Rank Matrix Recovery
- Structured Low-Rank Recovery of Piecewise Constant Signals with Performance Guarantees
- Acceleratating two-dimensional infrared spectroscopy while preserving lineshapes using GIRAF
- Calibration-free B0 correction of EPI data using structured low rank matrix recovery
- Recovery of damped exponentials using structured low rank matrix completion
- Super-Resolution MRI Using Finite Rate of Innovation Curves
- A Generalized Structured Low-Rank Matrix Completion Algorithm for MR Image Recovery
Union of Surfaces model for data living on manifolds
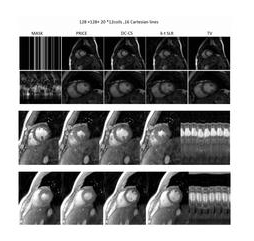
Current cardiac MR imaging exams are long (> 1hr) and require the subject to hold their breath several times. These long scans are often challenging for several patient populations (e.g. obese subjects and patients with compromised pulmonary function). Our main goal is to develop a short 3-D free-breathing & un-gated cardiac imaging protocol to evaluate cardiac structure, function, perfusion, and fibrosis in obese subjects in a short scan time.
We model the time profiles as points living on a manifold. We introduce a union of surfaces model to represent such signals efficiently. We rely on a non-linear lifting or mapping of the data, which will transform the union of surfaces representation to the well-know union of subspaces model. Relying on this connection, we introduce sampling theorems and fast algorithms for the recovery of signals with extensive non-linear structure.
Relevant papers
- Sampling of Planar Curves: Theory and Fast Algorithms
- Manifold recovery using kernel low-rank regularization: application to dynamic imaging
- Free-breathing & ungated cardiac MRI using iterative SToRM (i-SToRM)
- Accelerated Dynamic MRI Using Patch Regularization for Implicit Motion Compensation,
- Iterative Shrinkage Algorithm for Patch-Smoothness Regularized Medical Image Recovery,
- Nonlocal regularization of inverse problems: a unified variational framework
Software:
Learned Image Representations for Multidimensional Imaging
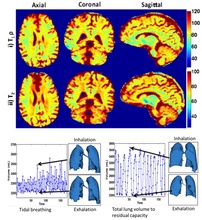
The recovery of the image data from sub-Nyquist sampled measurements using constrained image models is emerging as a promising approach to accelerate imaging. A challenge in using apriori selected models (eg. Fourier series, sparse wavelet representation) is that such models may not be efficient in representing the signal at hand. Our main focus is to develop novel theoretical framework and efficient algorithms to learn image representations from under-sampled measurements. The representations (blind compressed sensing, blind linear model termed as k-t SLR) enabled several accelerated MR imaging schemes. The proposed adaptive framework is a significant departure from classical approaches of using pre-determined dictionaries. We have also introduced theoretical guarantees for the recovery of such signals from undersampled data. The proposed framework is demonstrated to outperform classical compressed sensing methods in a range of applications including cardiac CINE MRI, myocardial perfusion MRI, dynamic lung imaging, and multi-parameter mapping where T1rho and T2 is simultaneously measured.
Relevant papers:
- Accelerated Dynamic MRI Exploiting Sparsity and Low-Rank Structure: k-t SLR
- A Fast Majorize-Minimize Algorithm for the Recovery of Sparse and Low Rank Matrices
- Accelerating Free Breathing Myocardial Perfusion MRI Using Multi Coil Radial k-t SLR
- Blind Compressive Sensing Dynamic MRI; Subspace Aware Recovery of Low Rank and Jointly Sparse Signals
- Blind Compressed Sensing Enables 3D Dynamic Free Breathing MR Imaging of Lung Volumes and Diaphragm Motion
Software:
MATLAB codes for blind compressed sensing (BCS) dynamic MRI; Matlab codes for k-t SLR
High Resolution Metabolic MRI using Learned Models
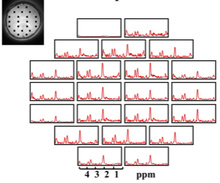
MRI provides several contrast mechanisms to examine the metabolism and tissue microstructure. There are certain challenges in MRI: the low signal to noise ratio of these imaging schemes, the long scan time, and the vulnerability to several artifacts. CBIG introduces novel pulse sequences and image reconstruction algorithms to mitigate the aforementioned problems. The learned models are observed to be highly effective in suppressing the artifacts while significantly enhancing spatial resolution and quality.
Relevant Papers:
- Compartmentalized low-rank recovery for high resolution lipid unsuppressed MRSI
- Sparse Spectral Deconvolution Algorithm for Noncartesian MR Spectroscopic Imaging
- Robust Reconstruction of MRSI Data Using a Sparse Spectral Model and High Resolution MRI Priors
- High-Resolution MRS in the Presence of Field Inhomogeneity via Intermolecular Double-Quantum Coherences on a 3-T Whole-Body Scanner
- Compartmentalized Low-Rank Regularization with Orthogonality Constraints for High-Resolution MRSI
- Improved Model-Based Magnetic Resonance Spectroscopic Imaging
High-Resolution Brain Connectivity Mapping
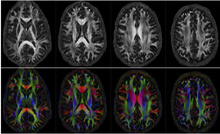
The imaging of water diffusion using MRI enables the imaging of axonal brain connectivity. CBIG's focus in this research is to understand the higher signal-to-noise ratio (SNR) offered by the ultra-high field (UHF) strengths to map brain connectivity at unprecedented spatial and angular resolutions. We utilize novel algorithms to minimize inter-shot motion induced distortions in multi-shot MRI; the multi-shot acquisition scheme provides improved SNR, which translates to higher spatial resolution. We also introduce novel compressed sensing strategies and joint K-Q undersampling strategies to further improve resolution.
Relevant Papers:
- Multi-Shot Sensitivity-Encoded Diffusion Data Recovery using Structured Low-Rank Matrix Completion (Mussels)
- Comprehensive Reconstruction of Multi-Shot Multi-Channel Diffusion Data using Mussels
- Acceleration of High Angular and Spatial Resolution Diffusion Imaging using Compressed Sensing with Multi-Channel Spiral Data
- Fast Iterative Algorithm for the Reconstruction of Multi-Shot non-Cartesian Diffusion Data
Fast Graph Search Algorithms for Fat-Water MRI

The decomposition of MRI data to generate water and fat images has several applications in medical imaging. CBIG introduces a novel algorithm to overcome the problems associated with current analytical and iterative decomposition schemes, such as lack of flexibility and convergence to local minima. We introduce a rapid graph, search based algorithm, known as GOOSE, which is robust in fat-water swaps in challenging applications. GOOSE has worked with great success, improving the state of fat-water MRI examination significantly.
Relevant Papers:
- Fat water decomposition using GlObally Optimal Surface Estimation (GOOSE) algorithm
- Algebraic decomposition of fat and water in MRI, 3D GlObally Optimal Surface estimation (3D-GOOSE) algorithm for fat and water separation
Software:
GlObally Optimal Surface Estimation (GOOSE) for fat water decomposition